Advanced usage¶
Making gene interval files¶
In order to work with other builds of human and mouse genome, or with
other organisms, the mk_pegs_intervals utility should be used to
generate a reference gene interval data file for input into the PEGS
analysis:
mk_pegs_intervals REFGENE_FILE
where REFGENE_FILE is the refGene annotation data for the genome
of interest.
These annotations can be obtained from the UCSC table browser as follows:
Select the genome and build of interest in the table browser
Ensure that NCBI RefSeq track is selected
Select get output to download the annotation to a local file.
An example of the table browser page for mm10 is shown below:
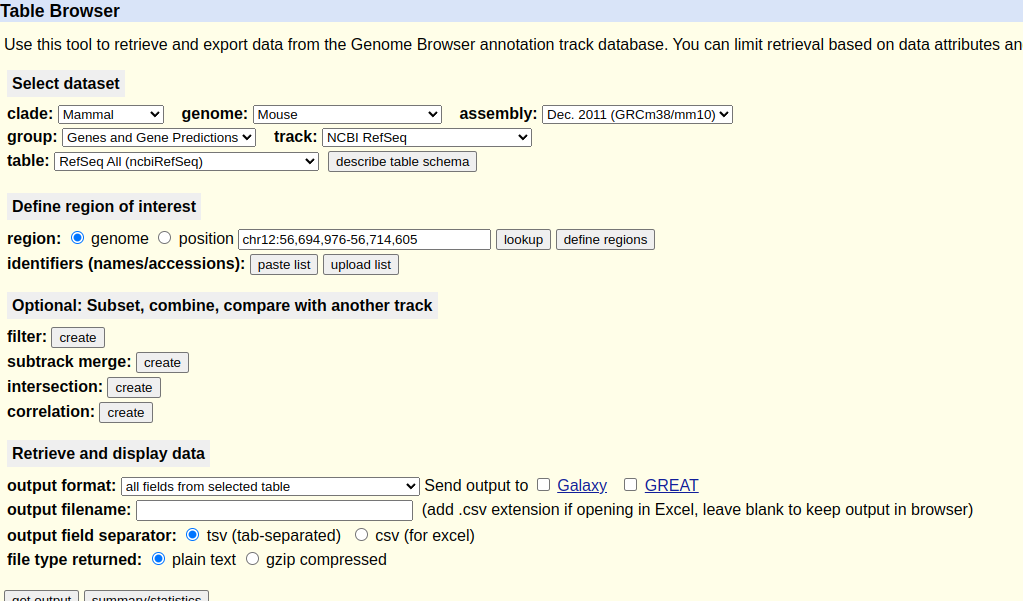
By default the output gene interval file will be called
<REFGENE_FILE>_intervals.bed; you can explicitly specify the
name using the -o option:
mk_pegs_intervals refGene_mm10.txt -o refGene_mm10_120719_intervals.bed
Customising the heatmap¶
Specifying the heatmap image format (PNG, SVG, PDF etc)¶
By default the heatmap is output from PEGS in PNG format, however
it is possible to change this in one of two ways, either:
Use the
--formatoption to explicitly specify the format aspng,svg,pdfetc, orUse the
-moption to explicitly specify the name for the output heatmap and use the appropriate file extension for the image format that you want (for examplemy_heatmap.svgwill automatically generate the heatmap as an SVG image).
Setting the heatmap axis labels¶
The default axis labels in the heatmap can be changed using the
--x-label and --y-label options, which specify the label
for the gene clusters (X-axis) and for the peak sets/distances
(Y-axis).
For example:
--x-label "Gene clusters" --y-label "Peaks and distances"
Changing the heatmap colours¶
The heatmap colour palette is the default Seaborn cubehelix palette
as generated by the seaborn.cubehelix_palette function:
A custom palette can be specified by using the --color option
to supply a base colour, for example:
--color seagreen
which produces a heatmap of the form:
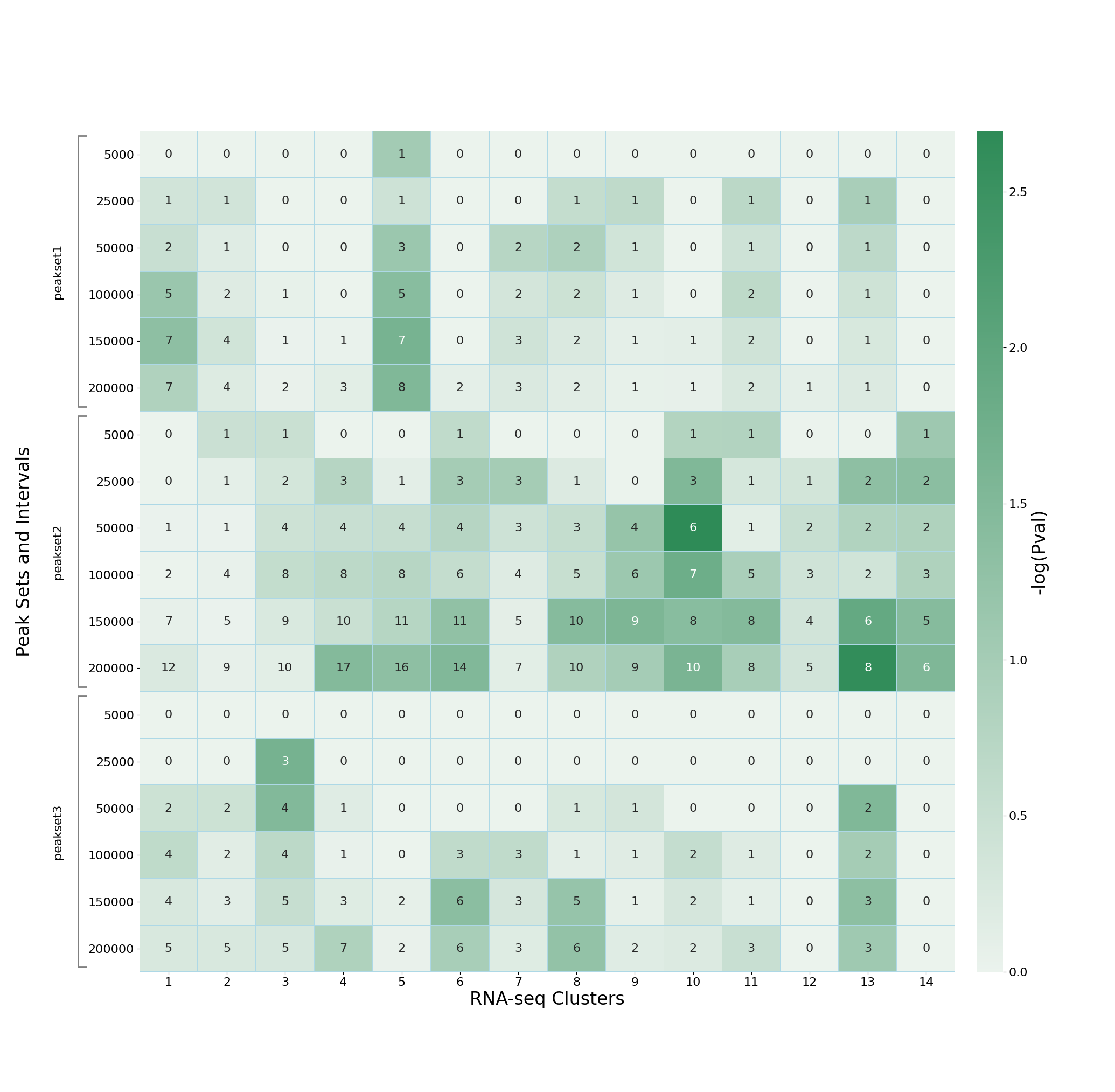
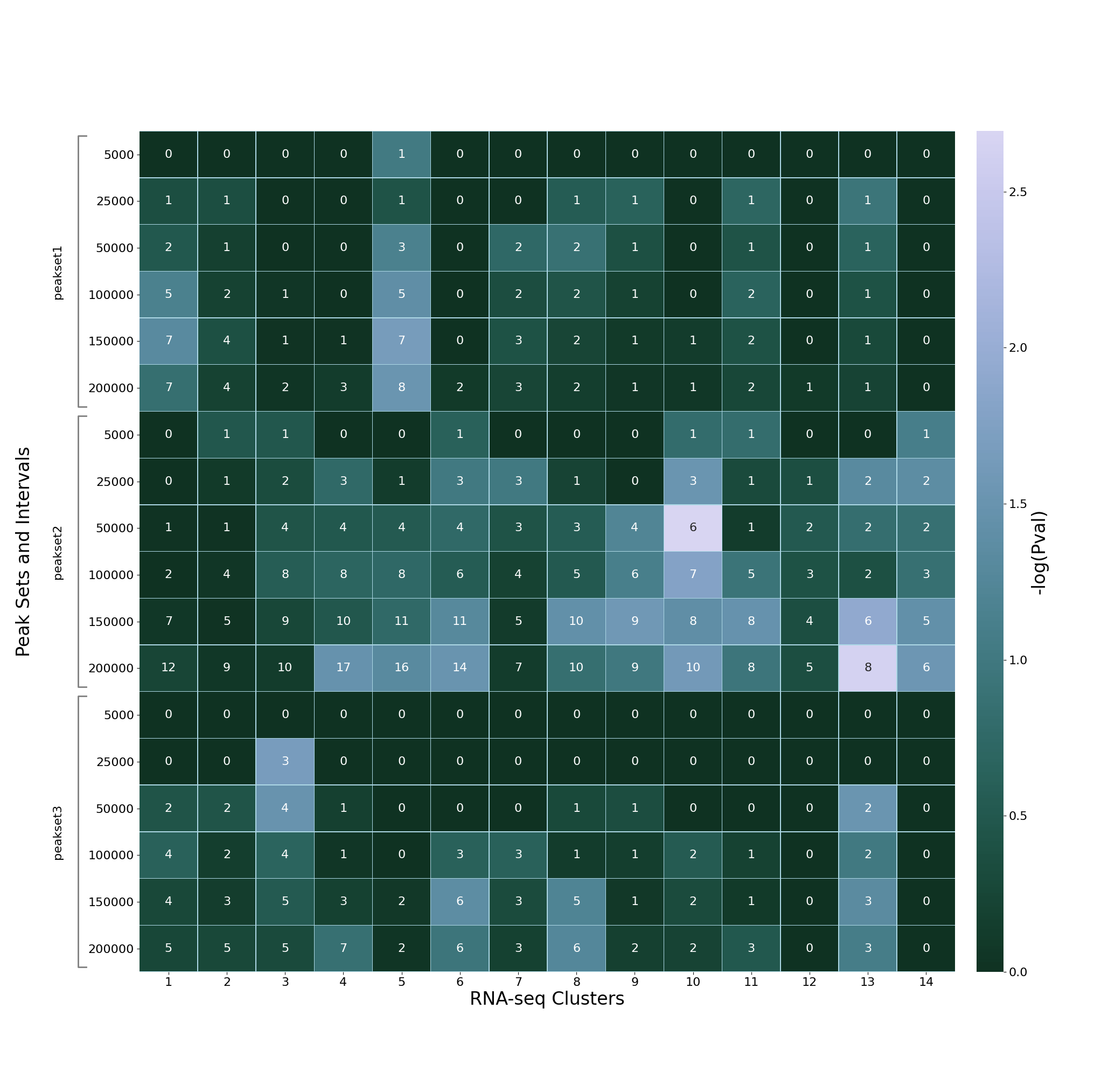
Alternatively the palette can be fully specified by setting the
parameters supplied to seaborn.cubehelix_palette, by using the
--heatmap-palette option. For example, specifying:
--heatmap-palette start=2 reverse=True
results in a “blue/green” heatmap (start=2) with low values
rendered as darker and high values as lighter (reverse=True):
Some other examples can be found at https://seaborn.pydata.org/tutorial/color_palettes.html#sequential-cubehelix-palettes